Coordinator
- Javier Paz-Ares, Centro Nacional de Biotecnología, C.S.I.C., Madrid.
Subproject Principal Investigator
- José Luis Micol, Instituto de Bioingeniería, Universidad Miguel Hernández.
Subproject Investigator:
- María Rosa Ponce, Instituto de Bioingeniería, Universidad Miguel Hernández.
Integrated project on Arabidopsis functional genomics
Most fields of the Biology of our century are suffering radical changes caused by the rapid spreading of the methods of Genomics, the large-scale analysis of gene structure and function. Such a methodological and conceptual renovation is only comparable to that of the Industrial Revolution in the late 18th century, when extensive mechanization of production systems resulted in a shift from home-based hand manufacturing to large-scale factory production, and involves radical changes in the experimental approaches, in the nature of the instruments used as tools and in the scale of the research tasks to be undertaken. These changes will not effectively occur without profound modifications in the way of thinking of scientists as well as in the behaviour of communities sharing scientific goals. Aware of this, the research teams making the present proposal aim to implement new national structures, resources and services allowing the use of genomic approaches to basic and applied research in plant biology. To this end, we propose several high-throughput and large-scale initiatives, using as experimental organism Arabidopsis thaliana, the par excellence model system in plant biology. In the study of this plant species are focused several excellent groups in Spain, whose dynamism and demonstrated performance make them an ideal community to test an organizational model for scientific activities unprecedented in our country, a model that might be used later for other scientific communities. We specifically propose the following activities in seven fields, all of which converge with, complement and reinforce recent international trends observed in current programs of Genomics developed in foreign countries. 1) Transcriptome analysis, by means of the production and use of DNA chips; 2) Análisis of the phosphoproteome and nuclear proteome; 3) Isolation of null mutants by insertional mutagenesis, including the identification of genomic sequences flanking the insertion; 4) Production of allelic series for specific genes, including point mutations and deletions; 5) Automated high resolution gene mapping, to facilitate the positional cloning of novel genes from their untagged mutant alleles; 6) Identification and cloning of open reading frames in chromosome 4; and 7) Exploitation of natural variability, by means of the collection and characterization of natural races in Spain and filogenomic studies to identify conserved genomic regions involved in the regulation of gene expression. The above mentioned seven initiatives will be implemented in seven facilities, which will serve themselves and to all the remaining groups that endorse this proposal.
Subproject: Automated gene mapping
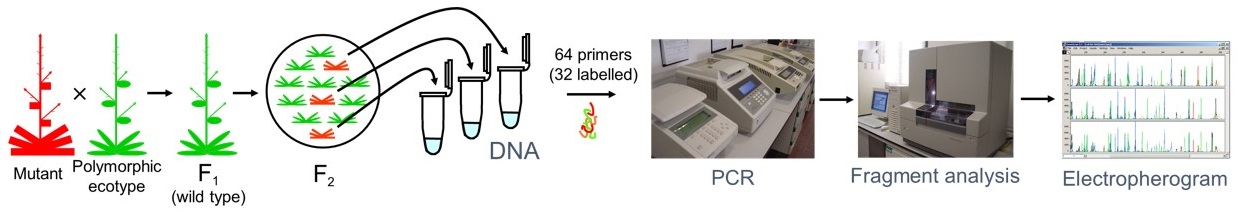
The first goal of genetic approaches to the study of gene function is its identification from the isolation of mutants. In Arabidopsis thaliana, the isolation of mutants tagged by insertional mutagenesis is assumed to be instrumental to rapidly identify the flanking genomic sequences, which correspond to the gene carrying the mutation. There is a number of drawbacks, however, in the use of the otherwise most efficient insertional mutagen in Arabidopsis thaliana, the T-DNA segment of the Ti plasmid from Agrobacterium tumefaciens. Indeed, T-DNA in many cases generates multiple unlinked insertions or internally reorganized and large concatameres and in some instances causes untagged mutations due to abortive insertions. Such a peculiar behavior of T-DNA based vectors frequently prevents the cloning of the genes damaged by their insertions. In addition, most T-DNA induced mutations are null and many cause recessive lethality.
The use of physical or chemical mutagens represents an alternative to insertional mutagenesis. A major objection usually raised to the use of these mutagens is that positional approaches must be followed for the cloning of the corresponding genes. Although until recently map-based cloning has been considered time-consuming and expensive in Arabidopsis thaliana, techniques developed in recent years, and the information provided by the Arabidopsis Genome Initiative, now make it possible to complete positional cloning projects in a short period of time. In addition, physical and chemical mutagens are more efficient than T-DNA and induce null and lethal alleles as well as hypomorphic and viable alleles.
The specific objective of this subproject is the implementation of a facility of automated, high-resolution gene mapping of untagged Arabidopsis thaliana mutants. Such a facility will use samples received from the research groups that endorse this proposal, consisting in at least 50 F2 individuals from an outcross involving a homozygous plant for a mutation of interest. This plant material will be used to extract DNA and perform a SSLP linkage analysis that will allow a low-resolution mapping yielding a rough map position within a 15 cM interval. In a second step, DNA from 450 additional F2 individuals will be extracted to make possible a high resolution SSLP linkage analysis intended to identify a contig of two or three BAC clones containing the gene affected in the mutant under study.